User guide - How to use OGP
Statistical analysis module provides an overview of the database status: number of recorded proteins, sites and site-specific glycans; distribution of recorded species and pathway analysis or disease clustering for human source recorded O-glycosylated proteins.
Database search module is designed for users to search with parameters like uniprot protein accession number, protein name, species, sample source or published year or peptide sequence. So far, it allows users to search O-glycoproteins with accession number, protein name, gene and glycan structure. It also allows users to directly linked to Uniprot for registered proteins and to NCBI PubMed for related reference for specific proteins or sites.
1. Users can search glycoproteins by accession number, protein name, gene name or single glycan structure (e.g. GalNAc;HexNAc;NeuAcHexHexNAc).
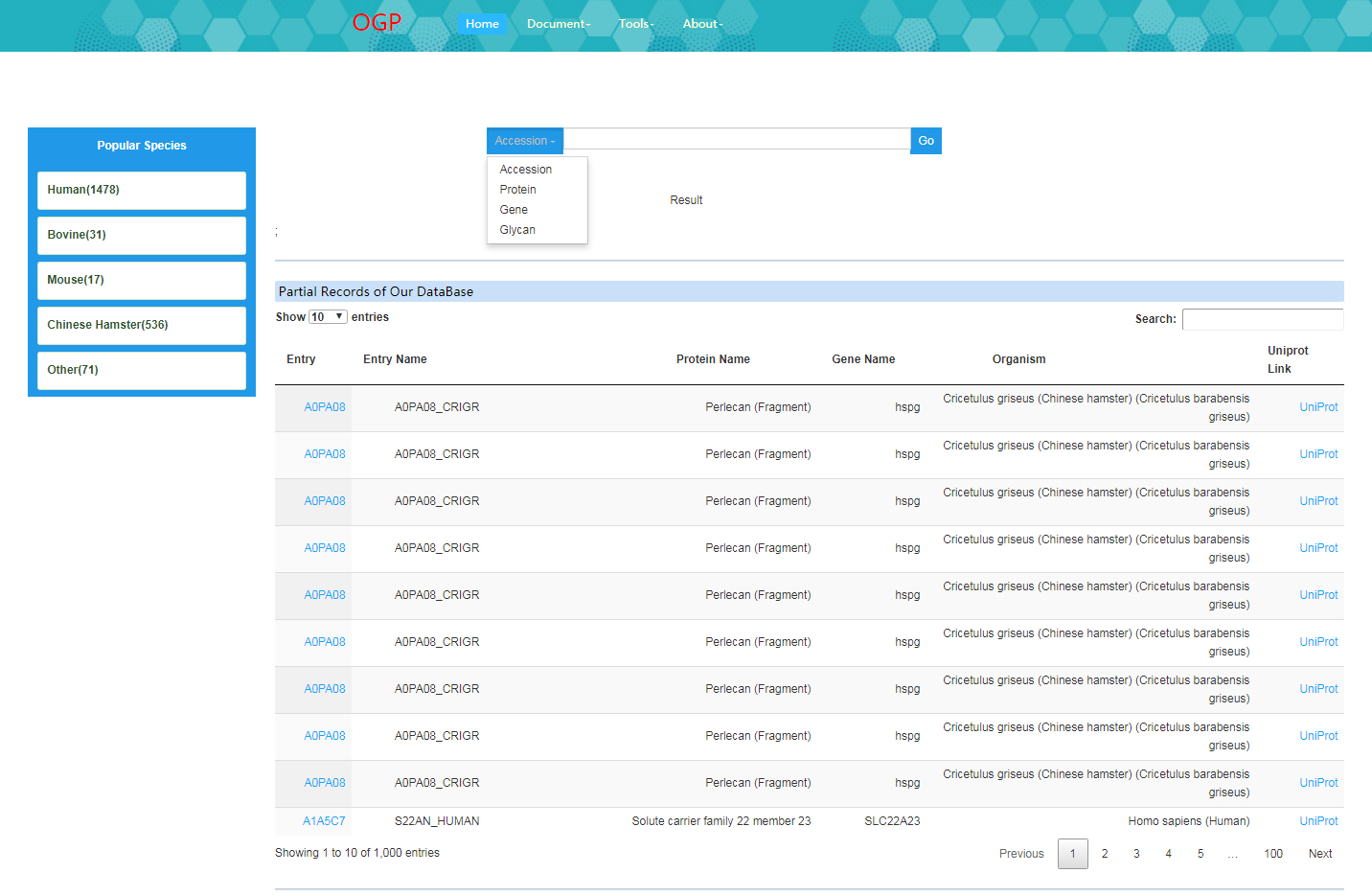
2. The query results include protein information, overview of O-glycosylation on protein level, O-glycosylated status with site specific glycans and techniques used (e.g. enzymes or instruments) and related literatures.
If there are multiple records response to one search, the related records will be displayed on top of the page.
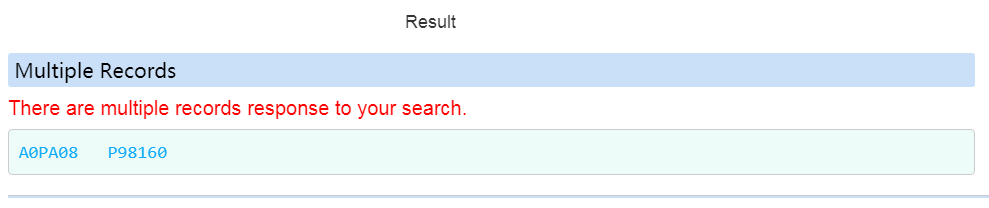
Overview
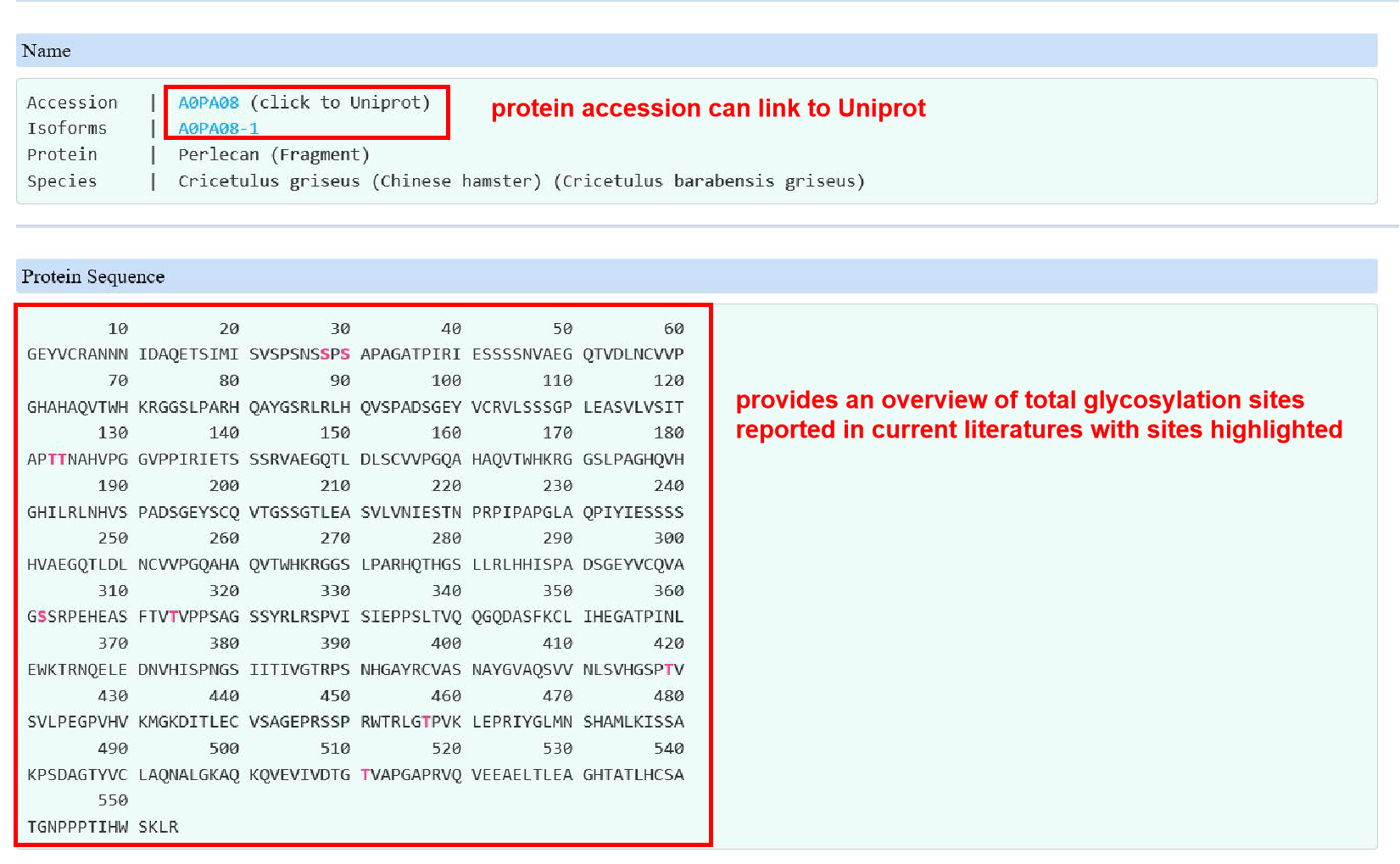
Sites and glycans
Methods
Corresponding references
Site prediction module includes an inherent example of site prediction modeling and a button where users can submit ordered sequence (11 amino acid with S/T in the middle of the sequence) of fasta-type protein sequence to get site prediction results.
1. On glycosite prediction page, two methods are provided: by uploading sequence aligned as the example file with targeted sites(S/T) centered or simple submit a protein sequence in fasta format.
2. Click "predict" to generate prediction results. The results will be shown on the right column. You can also click download to figure out which sites are likely or unlikely to be O-glycosylated.
Data submit module allows users to comment as well as summit high confident O-glycosylation data that consists well annotated mass spectral with clear site determination evidence or fragment ions to make this database more comprehensive.
We will be appreciate to receive suggestions or data from users. In the data summit page, we provide two ways to submit new discoveries: by uploading file in the format of example file (available on this page) or simply filling the form.
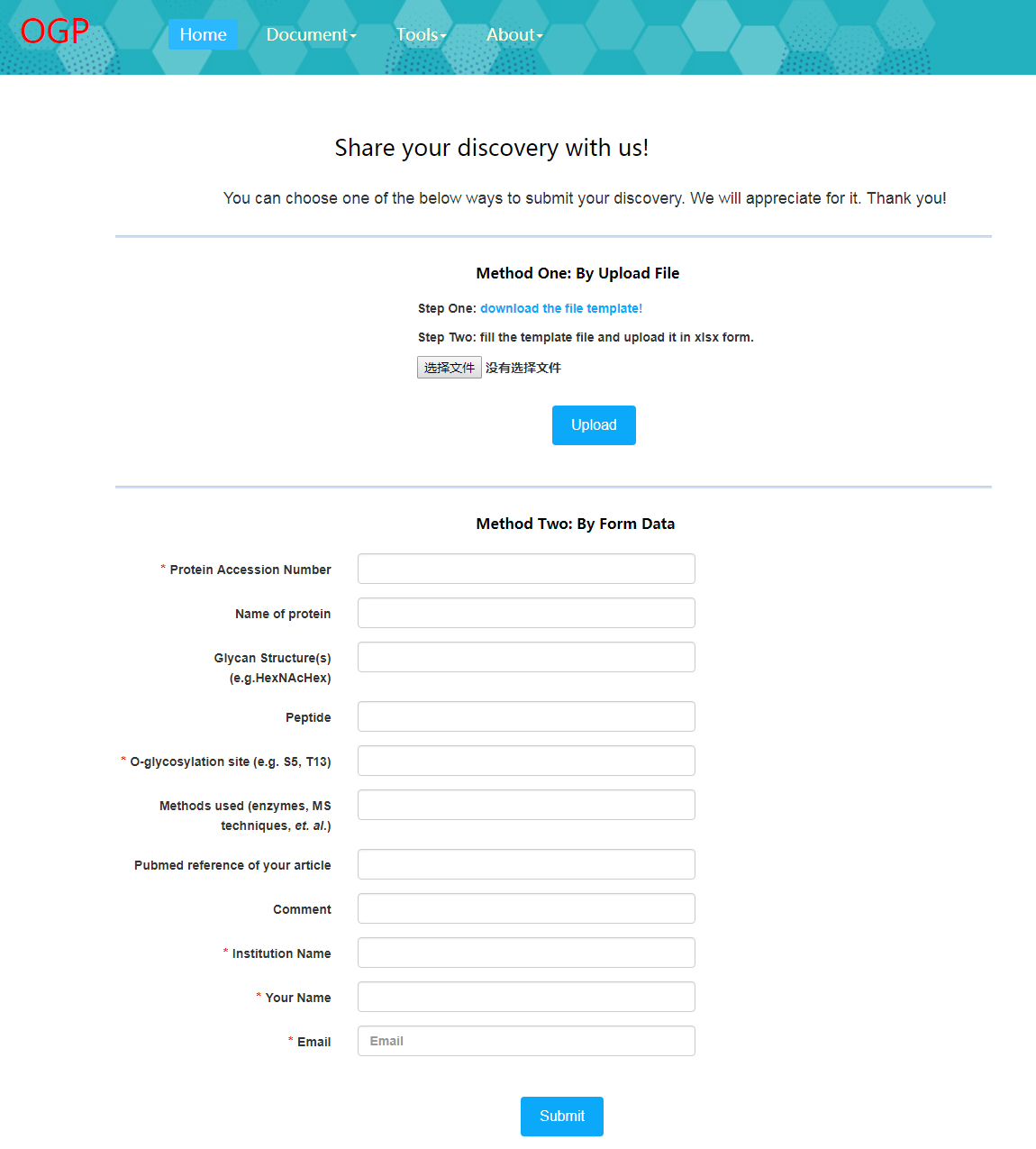
When user uploads the data by file, there will be a real time feedback shown below.
Download modules allow users to download our O-glycosylation data for research.
Copyright © 2017 School of Life Sciences Fudan University All Rights Reserved. 沪ICP备17021429号
Adress:Dong An Road, Fenglin Compus, Fudan University
Email: wqcao@fudan.edu.cn; weiqiancao09@fudan.edu.cn